Introduction
PyLiPD is a Python package for handling (LiPD) datasets.
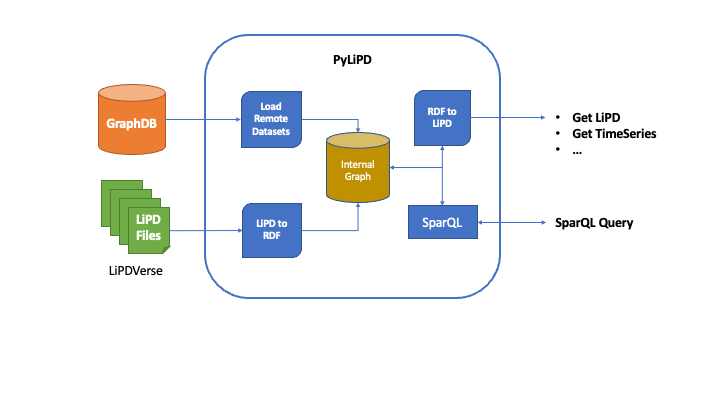
PyLiPD loads the Linked Paleo Data (LiPD), either locally or online, and converts them internally into (RDF graphs) for further querying. Alternatively, PyLiPD can also read an RDF Knowledge Base like GraphDB directly that is populated by the LiPD datasets converted into RDF graphs. In short, it allows you to work seamlessly with LiPD files stored on your computer, on the web or in a dedicated database meant to work with graphs.
PyLiPD can use the internal/remote graph representation to either answer (SparQL) queries about the datasets, or simply convert it back to LiPD, or get the TimeSeries objects across multiple datasets for further analysis by packages such as (Pyleoclim).
The package makes working with the Graph representation easier. You do no need to learn SparQL to start working with PyLiPD. However, SparQL is a fast, efficient language that makes querying much easier so we won’t stop you from learning it.
Getting Started
Working with PyLiPD